gene_list = ['EGFR', 'KRAS', 'MYC', 'RB', 'TP53']
print(gene_list)['EGFR', 'KRAS', 'MYC', 'RB', 'TP53']Students will be able to use data structures and control flows to make algorithms.
Data structures are basically just that - they are structures which can hold some data together. In other words, they are used to store a collection of related data. These are particularly helpful when working with experimental data sets. There are four built-in data structures in Python - list, tuple, dictionary and set.
There are four built-in data structures in python: lists,tuples, sets, and dictionary. In this lesson, we will learn about each data structures.
A list is a data structure that holds an ordered collection of items i.e. you can store a sequence of items in a list. This is easy to imagine if you can think of a shopping list where you have a list of items to buy, except that you probably have each item on a separate line in your shopping list whereas in Python you put commas in between them.
Here are important properties of lists:
Let’s create a list containing five different genes. In Python, a list is created by placing elements inside square brackets [], separated by commas.
Now let’s learn what we can do with lists.
We can access list items using index opreator. In python, indices start at 0.

Let’s try to access the second gene in our gene_list.
Python also has ‘negative indices’ which can be very convinient when we want to access last i-th item. Last item in the list can be accessed with index of -1. Let’s try to access the second to last item in gene_list.
We can also access a range of items in al list using the slicing operator :. Let’s try to access the first three items in gene_list. Note that the start index is inclusive, but end index is exclusive. We will see later when this can be useful.
# first three items
print(gene_list[0:3])
# beginning index (0) can be ommitteed
print(gene_list[:3])['EGFR', 'KRAS', 'MYC']
['EGFR', 'KRAS', 'MYC']What should we do if we want to access the last three items?
Now, we are interested in a new cancer gene, PTEN, and want to add this to our gene list. How should we do this? There are a few ways to do this, and one way is using a list method called, append.
Oop, we were supposed add BRAF instead of PTEN! What should we do? Since lists are mutable, we can repalce PTEN with BRAF.
There are other useful list methods that we can use to alterate and describe lists.
list.append(x): Add an item x to the end of the list.
list.remove(x): Remove the first item from the list whose value is equal to x.
list.count(x): Return the number of times x appears in the list.
reversed(l): Reverse the order of items in list l and return the new list.
sorted(l): Sort the order of values in list l (default is in ascending order) and return the new list.
len(l): Return the length of list l
Tuples are ordered collection of values. Tuples are similar to lists with one major difference. They are are immutable, meaning that we cannot change, add, or remove items after they are created. We an create a tuple by placing comma-seperated values inside ().
Similarly to lists, we can use indexing and slicing to access items
MYC
('RB', 'TP53')Can we replace KRAS with NRAS for tuples?
--------------------------------------------------------------------------- TypeError Traceback (most recent call last) Cell In[11], line 1 ----> 1 gene_tuple[1] = 'NRAS' TypeError: 'tuple' object does not support item assignment
This raises error because tuples are immutable.
Python set is an unordered collection of unique items. They are commonly used for computing mathematical operations such as union, intersection, difference, and symmetric difference.
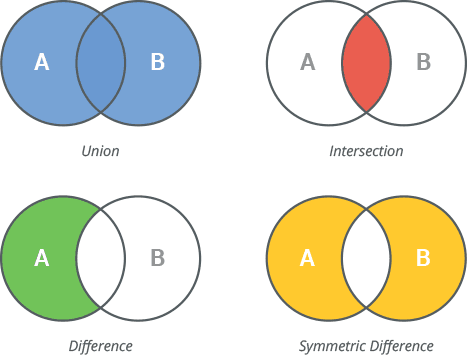
The important properties of Python sets are as follows:
Sets can be created by placing items comma-seperated values inside {}
We have upregulated genes in tumor tissues compared to normal tissues from two patients. We would like to know if there is a shared upregulated genes.
A dictionary is like an address-book where you can find the address or contact details of a person by knowing only his/her name i.e. we associate keys (name) with values (details). Note that the key must be unique just like you cannot find out the correct information if you have two persons with the exact same name.
Note that you can use only immutable objects (like strings) for the keys of a dictionary but you can use either immutable or mutable objects for the values of the dictionary.
Pairs of keys and values are specified in a dictionary by using the notation d = {key1 : value1, key2 : value2 }. Notice that the key-value pairs are separated by a colon and the pairs are separated themselves by commas and all this is enclosed in a pair of curly braces.
The following example of a dictionary might be useful if you wanted to keep track of ages of patients in a clinical trial.
agesDict = {'Karen P.' : 53, 'Jessica M.': 47, 'David G.' : 45, 'Susan K.' : 57, 'Eric O.' : 50}
print(agesDict){'Karen P.': 53, 'Jessica M.': 47, 'David G.': 45, 'Susan K.': 57, 'Eric O.': 50}We can access a person’s age (value) using his/her name (key). Let’s find out Eric O.’s age.
A new patient is enrolled into the clinical trial. Her name is Hannah H. and her age is 39. We can add a new item to the dictoary.
{'Karen P.': 53, 'Jessica M.': 47, 'David G.': 45, 'Susan K.': 57, 'Eric O.': 50, 'Hannah H.': 39}Now we have learned the four basic python data structures: list, tuple, set, and dictionary. We have jsut touched the surface of these data structures. To learn more about these data structures and how to use them, please refer to the references!
The following exercises will help you better understand data structures.
num_listmed_dict >* Lisinopril: 23.07 >* Gabapentin: 86.27 >* Sildenafil: 169.94 >* Amoxicillin: 17.76 >* Prednisone: 13.81med_dict to calculate how much it will cost if a patien tis treated with Lisinopril and Prednisone.Decision making is required when we want to execute a code only if a certain condition is satisfied.
The if…elif…else statement is used in Python for decision making. We can use these statments to execute a block of code only when the condition is true. The if…elif…else statement follows this syntax. Note, elif is abbreviation for else if.
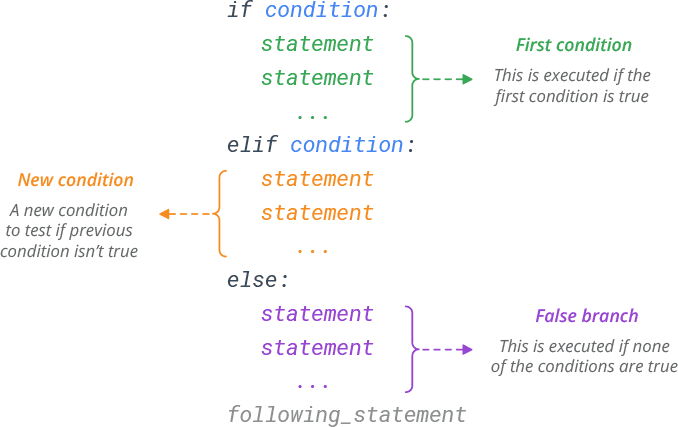
Let’s think of a dose-finding clinical trial. We first treat three patients with dose x. iIf no patients shows toxic side effects, we increase the dose. If one patient shows toxicity, we treat another three patients to learn more. If more than one patients show toxicity, we stop at that dose. Let’s make this into python code. You can change the value of n_toxic to see how the script works.
There are two types of loops in python: while loops and for loops. Loops are useful when we want to performe the same task repetitively.
A while loop is used when you want to perform a task indefinitely, until a particular condition is met. For instance, we want to enroll new patients to a clinical trial until we have 30 patients.
enrolled patient 1
enrolled patient 2
enrolled patient 3
enrolled patient 4
enrolled patient 5
enrolled patient 6
enrolled patient 7
enrolled patient 8
enrolled patient 9
enrolled patient 10
enrolled patient 11
enrolled patient 12
enrolled patient 13
enrolled patient 14
enrolled patient 15
enrolled patient 16
enrolled patient 17
enrolled patient 18
enrolled patient 19
enrolled patient 20
enrolled patient 21
enrolled patient 22
enrolled patient 23
enrolled patient 24
enrolled patient 25
enrolled patient 26
enrolled patient 27
enrolled patient 28
enrolled patient 29
enrolled patient 30What do you think will hapen if we use while True?
For loops are used for iterating over a sequence of objects i.e. go through each item in a sequence. Iterable of items include lists, tuples, dictonaries, sets, and strings. For loop has the following syntax:
Let’s look at an example of a for loop.
We can also use the range function to do the same thing. range function takes three parameters: start(default 0), end, and steps (default 1). Like slicing, the start is inclusive but the end is exclusive.
Before going into exercise, we need to learn a handy operation +=.
is equivalent to
for and if.Create two new variables, comp_oligo1 and comp_oligo2, that are the complementary DNA sequences of oligo1 and oligo2 (hint: A <-> T and G <-> C)
Cell In[32], line 4 if nuc == #something: ^ SyntaxError: invalid syntax
dose_list = [1, 2, 3, 5, 8, 13]. We want to increase the dose until we hit the maximal tolerated dose (MTD). For simplicity, we will increase dose when there is less than two patients out of three patients with toxicity and stop otherwise. The last dose before at least two patients have toxicity is declared MTD. We will look into the future and assume that we know how many patients will have toxicity at each dose tox_list = [0, 0, 1, 1, 2, 2]. Find the MTD using while.Hopefully, you can see how importing data from and exporting data to csv or other delimiter separated files will be helpful for research. Now it’s time to practice your skills with built-in data structures and data frames!
https://www.programiz.com/python-programming/list
https://python.swaroopch.com/data_structures.html
https://docs.python.org/3/tutorial/datastructures.html
https://www.learnbyexample.org/python-tuple/